class: center, middle, inverse, title-slide .title[ # Low-dimensional Representation of Dynamic Systems with Phase-lags ] .subtitle[ ## A Composition in Several Movements ] .author[ ### by Mendelssohn ] .date[ ### 2025-06-03 ] --- # Old School (not "state-of-the art") Mostly lack of fancy acronyms and buzzword titles + but will have lots of pretty pictures because it must be true then -- no ratios of probabilities that are on (-1, 1) scale -- meaning of scale does not change over time -- conditional and unconditional probabilities not the same -- Take into consideration spatial and temporal dependence -- Concerned with dynamics, not statics -- Stationary versus non-stationary -- global vs. local properties, stability of estimates -- large number of parameters - some sort of regularization or shrinkage --- class: inverse, center, middle # Things That Have Been "Conclusively Shown" - 4 Thoughts --- # El Nino's are more frequent, more intense  --- # El Nino's have the same signature  --- # Coastal Mid-latitude dynamics driven by tropics  --- # Stationarity (relatively short time-series)  --- # Why a Duck? --  --- ``` r # Evidence for simple phase shift in data require(xts) load('chla_modelts_results.RData') series_dates <- seq(as.Date('1997-9-16'), as.Date('2010-12-16'), by = 'month') temp_data <- modelts_results$seasons[1:5, 1, ] temp_data <- xts(t(temp_data), order.by = series_dates) p <- plot(temp_data, grid.ticks.on = 'years') print(p) ```  --- ``` r # Zoomed phase shift in data require(xts) temp_data <- xts(temp_data[13:48, ], order.by = series_dates[13:48]) p <- plot(temp_data) print(p) ```  --- # How to do ? -- EOFs !!!??!!?? -- Why? -- - mumble-mumble -- - mumble-mumble -- - VARIANCE! -- - mumble-mumble -- - ??? -- - It Works!! --- # Cycles and phases, quick review  --- # State-space representation of cycles $$ `\begin{equation} \left[ \begin{array}{c} \psi_{t} \\ \psi_{t}^{*} \end{array}\right] = \rho \left[ \begin{array}{rr} \cos \lambda_{c} & \sin \lambda_{c} \\ -\sin \lambda_{c} & \cos \lambda_{c} \end{array}\right] \left[ \begin{array}{c} \psi_{t-1} \\ \psi_{t-1}^{*} \end{array}\right] + \left[ \begin{array}{c} \kappa_{t} \\ \kappa_{t}^{*} \end{array}\right], \qquad t=1,\ldots,T, \end{equation}` $$ - Starting with `\(\psi_0 = a\)` and `\(\psi_0^* = b\)`. --- # Why Does That Work? - Deterministic cycle `\(\psi_t = a \cos(\lambda t -b )\)` - Amplitude `\(a\)`, Phase `\(b\)`, Frequency `\(\lambda\)` - rewritten as `\(\alpha \cos(\lambda t) + \beta \sin(\lambda t)\)` - `\(\alpha = a \cos b\)`, `\(\beta = a \sin b\)` - reverse, `\(a = \alpha^2 + \beta^2\)`, `\(b = \tan^{-1}(\beta / \alpha)\)` - Follows from identities: + `\(\cos(\lambda \pm \xi) = \cos \lambda \cos \xi \mp \sin \lambda \sin \xi\)` and + `\(\sin (\lambda \pm \xi) = \cos \lambda \sin \xi \pm \sin \lambda \cos \xi\)` - Essentially the series and its Hilbert transform --- # How is phase 0 determined if all series are only relative to each other? --- class: center, center  --- # Complex EOFs (CEOFs) - Take Hilbert transform of the data - Do usual EOF calculations except for complex-valued series and matrices: + calculate amplitudes from real-part as above - `\(\alpha^2 + \beta^2\)` + calculate phases from `\(\tan^{-1}\)` --- # Problems with CEOFs - Assumes stationarity -- - Shifts all frequencies at once - only makes sense over limited bandwidth + Requires filtering the series -- - Filtering and Hilbert transform require complete data -- - Implicitly estimating a ton of parameters + all weights are non-zero, for large number of series adds a lot of noise + essentially least-squres, a few outliers can overly influence results + lasso-type estimators for real-matrices could be used -- - Estimates are global in time and space + no regulariztion in time + no "smoothing" in space + local dynamics can be overwhelmed in the analysis --- # CEOF Python code ``` python import numpy as np from scipy.signal import hilbert pi = np.pi data = np.genfromtxt('soda_v_sea_cyc.csv', delimiter=',') ntim, npt = data.shape # Hilbert transform the data data = hilbert(data, axis = 0) # covariance matrix c = np.matmul(np.conj(np.transpose(data)), data) / ntim # CEOF lamda, loadings = np.linalg.eigh(c, UPLO = 'U') lamda = np.flip(lamda, axis = 0) loadings = np.fliplr(loadings)[:, 0:12] pcs = np.matmul(data, loadings) # Spatial amplitude and phase S = np.power(loadings * np.conj(loadings), 0.5) theta = np.arctan2(np.imag(loadings), np.real(loadings)) theta2 = theta * 180./pi # Temporal amplitude and phase Rt = np.power(pcs * np.conj(pcs), 0.5) phit = np.arctan2(np.imag(pcs), np.real(pcs)) phit2 = phit * 180./pi ``` --- # Using State-space models with CEOFs - Fills in missing-data - Different Components filter the data + provides a certain amount of temporal regularization - Univariate model estimated for each series at each location + A ton more parameters estimated - Used the component of interest in the CEOF --- # R State-space Helper Function - update_modeltsc ``` r update_modeltsc <- function(pars, model) { finite_test <- 0.5 * log(.000001) if (pars[1] < finite_test) { pars[1] <- finite_test } model$H[1,1,1] <- exp(2. * pars[1]) freq <- (2.*pi)/(2. + exp(pars[5])) mycycle <- SSMcycle(period = (1./freq), Q = matrix(NA)) mycycle$T[abs(mycycle$T < 1.0e-10)] <- 0. damp <- abs(pars[6])/sqrt(1 + pars[6]**2) temp1 <- 1 - damp**2 temp2 <- exp(2 * pars[4]) var_cycle <- temp2 * temp1 diag(model$Q[,,1]) <- exp(c(2 * pars[2], 2 * pars[3], rep(var_cycle,11))) model$T[13:14,13:14,1] <- damp*mycycle$T return(model) } ``` --- # R State-space Helper Function - update_modelts ``` r update_modelts <- function(pars, model) { h1 <- exp(2. * pars[1]) if (h1 < .000001) { pars[1] <- .5 * log(.000001) } model$H[1,1,1] <- exp(2. * pars[1]) diag(model$Q[,,1]) <- c(exp(2*pars[2]), rep(exp(2*pars[3]), 11)) return(model) } ``` --- # R State-space Helper Function - update_modelsc ``` r update_modelsc <- function(pars, model) { require(KFAS) h1 <- exp(2. * pars[1]) if (h1 < .000001) { pars[1] <- .5 * log(.000001) } model$H[1,1,1] <- exp(2. * pars[1]) freq <- (2.*pi)/(2. + exp(pars[4])) mycycle1 <- SSMcycle(period = (1./freq), Q = matrix(NA)) damp <- abs(pars[5])/sqrt(1 + pars[5]**2) temp1 <- 1 - damp**2 temp2 <- exp(2 * pars[3]) var_cycle <- temp2 * temp1 diag(model$Q[,,1]) <- c(rep(exp(2*pars[2]), 11), rep(var_cycle,2)) model$T[13:14, 13:14, 1] <- damp*mycycle1$T return(model) } ``` --- # State-Space initial values ``` r irreg_init <- 0.5 * log(1) level_init <- 0.5 * log(.1) season_init <- 0.5 * log(.1) cycle_init <- 0.5 * log(1.8) freq1Init <- 8 dampInit <- 2 damp <- abs(dampInit)/sqrt(1 + dampInit**2) cycle1_init <- cycle_init/sqrt(1 - damp**2) freq <- (2.*pi)/(2. + exp(freq1Init)) modeltsc_inits <- c(irreg_init, level_init, season_init, cycle1_init, freq1Init, dampInit) npar_tsc <- length(modeltsc_inits) modelsc_inits <- c(irreg_init, season_init, cycle1_init, freq1Init, dampInit) npar_sc <- length(modelsc_inits) modelts_inits <- c(irreg_init, level_init, season_init) npar_ts <- length(modelts_inits) ``` --- # Example state-space model fit ``` r for (ilon in 1:no_lon) { for (ilat in 1:no_lat) { tempData <- sst[ilon, ilat, ] first_good <- min(which(!is.na(tempData))) if (is.finite(first_good)) { sst_modelts <- SSModel(sstData ~ SSMtrend(degree = 1 , Q = list(NA)) + SSMseasonal(12, Q=NA, sea.type = "trigonometric"), H = matrix(NA)) sst_modelts$T[abs(sst_modelts$T < 1.0e-10)] <- 0. sst_modelts_Fit <- fitSSM(model = sst_modelts, inits = modelts_inits, updatefn = update_modelts, method = "BFGS") sst_modelts_smooth <- KFS(sst_modelts_Fit$model, filtering = "state", smoothing = "state") level <- signal(sst_modelts_smooth, states = 'level')$signal season <- signal(sst_modelts_smooth, states = 'season')$signal modelts_results$levels[ilon, ilat, first_good:no_periods] <- level modelts_results$seasons[ilon, ilat, first_good:no_periods] <- season ll <- logLik(sst_modelts_Fit$model) modelts_results$model_aic[ilon, ilat] <- (-2 * ll) + (2 * npar_sc) modelts_results$model_bic[ilon, ilat] <- (-2 * ll) + log(nobs) * npar_sc modelts_results$model_aicc[ilon, ilat] <- modelts_results$model_aic[ilon, ilat] + 2 * npar_sc * (npar_sc + 1) / (nobs - npar_sc - 1) } } } ``` ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  --- # SST CEOF Notes - When phase shifts taken into account, similar regions north/south are more similar to each other than neighboring regions. + This was noted for trends by Mendelssohn and Schwing -- - El Nino's are seen in temporal series, but not all El Ninos are alike. --- # Frequency Domain EOF - What if we don't feel proper to phase shift all frequencies the same. - Calculate Spectral Density Matrix at any frequency - Proceed as in CEOF. -- We have already separated by frequency - any guesses how? ---  ---  ---  ---  ---  ---  --- # Hadley Notes - See El Ninos, but different ones on different components in differing ways. -- - See the Mid-latitude structure as in Parrish et al. -- - The sequence of phases look like spherical harmonics, noted in Mendelssohn and Bessey. --- # Canonical Analysis of Multiple Time-series in Frequency Domain - Multiple Approaches - Use symmetric formulation in Brillinger -- - Find the roots of: + `\(f_{yy}^{-1/2}(\lambda) f_{yx}(\lambda) f_xx^{-1}(\lambda) f_{xy}(\lambda) f_{yy}^{-1/2}(\lambda)\)` -- - If `\(V(\lambda)\)` is the solution above, the two weighting matrices are then + for x: `\(f{xx}^{-1}(\lambda) f_{xy}(\lambda) f_{yy}^{-1/2}(\lambda) V(\lambda\)` + for y: `\(f_{yy}^{-1/2}(\lambda) V(\lambda)\)` --- - Issues - same as with CEOF + stationarity, complete data, global estimates, many parameters + everything way over-determined, need pseudo-inverses, can give funny results - assumes linear relationship - may just be modeling that there is a seasonal, no predictive info beyond that ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  --- # Representing a changing seasonal in trigonometric form - Have a cycle term for each fundamental frequency, that is: + `\(\psi_t = \sum_{j=1}^{[s/2]} \psi_{j, t}\)` + Frequency for each `\(\psi_{j, t}\)` is `\(2 \pi j / s\)` --- # Phase shift of the trigonometric cycle - For a given cycle, we can shift in time as: + `\(\psi_1 = a \cos((t - \xi) \lambda) + b \sin((t - \xi) \lambda)\)` + `\(-.5\pi \le \xi \le .5\pi\)` - Cycle is shifted `\(\xi\)` time units to the "right". - But given our state components, the same shift can be done as: + `\(\cos(\xi \lambda) \psi_t + \sin(\xi \lambda) \psi_t^*\)` - by taking a given cycle and putting + `\([\cos(\xi \lambda), \sin(\xi \lambda) ]\)` in the observation matrix. --- ``` r # Function will be used to shift series phase_vector <- function(phase){ shift_vector <- array(NA_real_, dim = 11) shift_vector[1] = cos(phase * (2*pi/12)) shift_vector[2] = sin(phase * (2*pi/12)) shift_vector[3] = cos(phase * (4*pi/12)) shift_vector[4] = sin(phase * (4*pi/12)) shift_vector[5] = cos(phase * (6*pi/12)) shift_vector[6] = sin(phase * (6*pi/12)) shift_vector[7] = cos(phase * (8*pi/12)) shift_vector[8] = sin(phase * (8*pi/12)) shift_vector[9] = cos(phase * (10*pi/12)) shift_vector[10] = sin(phase * (10*pi/12)) shift_vector[11] = cos(phase * (12*pi/12)) shift_vector } ``` --- ``` r # Calculating shifts of a single seasonal require(KFAS) test_data <- modelts_results$seasons[1, 1, 4:160] test_model <- SSModel(test_data ~ SSMseasonal(12, Q = 0.00819, sea.type = 'trigonometric'), H = 4.64e-09) test_model_smooth <- KFS(test_model, filtering = "state", smoothing = "state") alphahat <- test_model_smooth$alphahat no_per <- length(series_dates[4:160]) temp_data <- array(NA_real_, dim = c(no_per, 5)) temp_data[, 3] <- as.vector(signal(test_model_smooth, states = 'season')$signal) temp_data[, 1] <- alphahat[, 2:12] %*% phase_vector(-pi/2) temp_data[, 2] <- alphahat[, 2:12] %*% phase_vector(-pi/4) temp_data[, 4] <- alphahat[, 2:12] %*% phase_vector(pi/4) temp_data[, 5] <- alphahat[, 2:12] %*% phase_vector(pi/2) temp_data <- xts(temp_data, order.by = series_dates[4:160]) ``` --- ``` r # Runstler's method does indeed shift the seasonal p <- plot(temp_data, grid.ticks.on = 'years') print(p) ```  --- ``` r # Zoom seasonal shift p <- plot(temp_data[13:48, ], grid.ticks.on = 'years') print(p) ``` 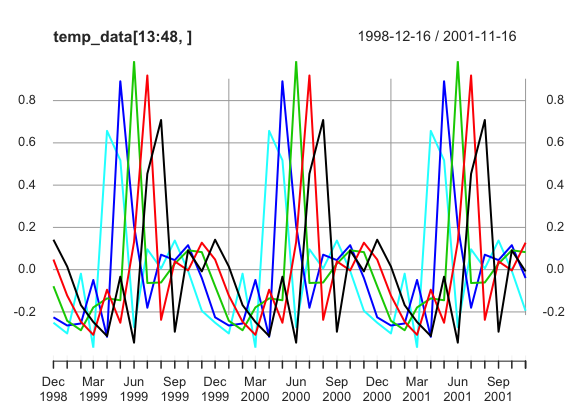 --- ``` r # Phase shift function redone to allow unconstrained parameter create_phased_z <- function(theta) { shift_vector <- array(NA_real_, dim = 11) freq <- 2*pi/12 e_shift <- (pi/freq) * (exp(theta)/(1 + exp(theta)) - 0.5 ) shift_vector[1] = cos(e_shift * freq) shift_vector[2] = sin(e_shift * freq) freq <- 4*pi/12 e_shift <- (pi/freq) * (exp(theta)/(1 + exp(theta)) - 0.5 ) shift_vector[3] = cos(e_shift * freq) shift_vector[4] = sin(e_shift * freq) freq <- 6*pi/12 e_shift <- (pi/freq) * (exp(theta)/(1 + exp(theta)) - 0.5 ) shift_vector[5] = cos(e_shift * freq) shift_vector[6] = sin(e_shift * freq) freq <- 8*pi/12 e_shift <- (pi/freq) * (exp(theta)/(1 + exp(theta)) - 0.5 ) shift_vector[7] = cos(e_shift * freq) shift_vector[8] = sin(e_shift * freq) freq <- 10*pi/12 e_shift <- (pi/freq) * (exp(theta)/(1 + exp(theta)) - 0.5 ) shift_vector[9] = cos(e_shift * freq) shift_vector[10] = sin(e_shift * freq) freq <- 12*pi/12 e_shift <- (pi/freq) * (exp(theta)/(1 + exp(theta)) - 0.5 ) shift_vector[11] = cos(e_shift * freq) shift_vector } ``` --- ``` r # update to parameters with phase shift update_model_seasphase <- function(pars, model, no_seasons) { len_pars <- length(pars) no_series <- dim(model$H)[1] h1 = exp(2. * pars[no_series]) finite_test <- 0.5 * log(.000001) pars_temp <- pars[1:no_series] pars_temp[pars_temp < finite_test] <- finite_test pars[1:no_series] <- pars_temp temp_H <- exp(2. * pars[1:no_series]) diag(model$H[, ,1]) <- temp_H diag(model$Q[2:12, 2:12, 1]) <- rep(exp(2. * pars[no_series + 1]), 11) phase_init <- pars[(len_pars - (2 * no_series) + 1):(len_pars - no_series)] phase_d_init <- pars[(len_pars - no_series + 1):len_pars] for (iseries in 1:no_series) { shift_vector <- create_phased_z(phase_init[iseries]) model$Z[iseries, 2:12, 1] <- phase_d_init[iseries] * shift_vector } return(model) } ``` --- ``` r # setting up KFAS for phase shift model - 100 series no_series <- dim(temp_data)[2] junk <- SSMtrend(degree = 1 , Q = 0., P1 = 0, P1inf = 0, a1 = 0.) junk1 <- SSMseasonal(12, Q = NA, sea.type = 'trigonometric', type = 'common', P1inf = diag(0, 11, 11), P1 = diag(10, 11, 11)) Q <- array(0., dim = c(11, 11, 1)) diag(Q[, , 1]) <- NA Z <- array(0, dim = c(no_series, 11, 1)) shift_vector <- create_phased_z(0.) Z[1, 1:11, 1] <- junk1$Z for (i in 1:no_series) { Z[i, 1:11, 1] <- shift_vector } T <- junk1$T H <- array(0, dim = c(no_series, no_series, 1)) diag(H[, , 1]) <- NA P1 <- array(0, dim = c(11, 11)) diag(P1) <- 10. P1inf <- array(0, dim = c(11, 11)) R = diag(1, 11, 11) R <- array(R, dim = c(11, 11, 1)) a1 <- array(0, dim = c(11, 1)) test <- SSModel(temp_data ~ SSMtrend(degree = 1 , Q = 0., P1 = 0, P1inf = 0, a1 = 0., type = 'common') + SSMcustom(Z, T, R, Q, a1, P1, P1inf), H = H) season_init <- 0.5 * log(.1) irreg_init <- 0.5 * log(.00001) phase_init <- 0. phase_d_init <- 1. ``` --- 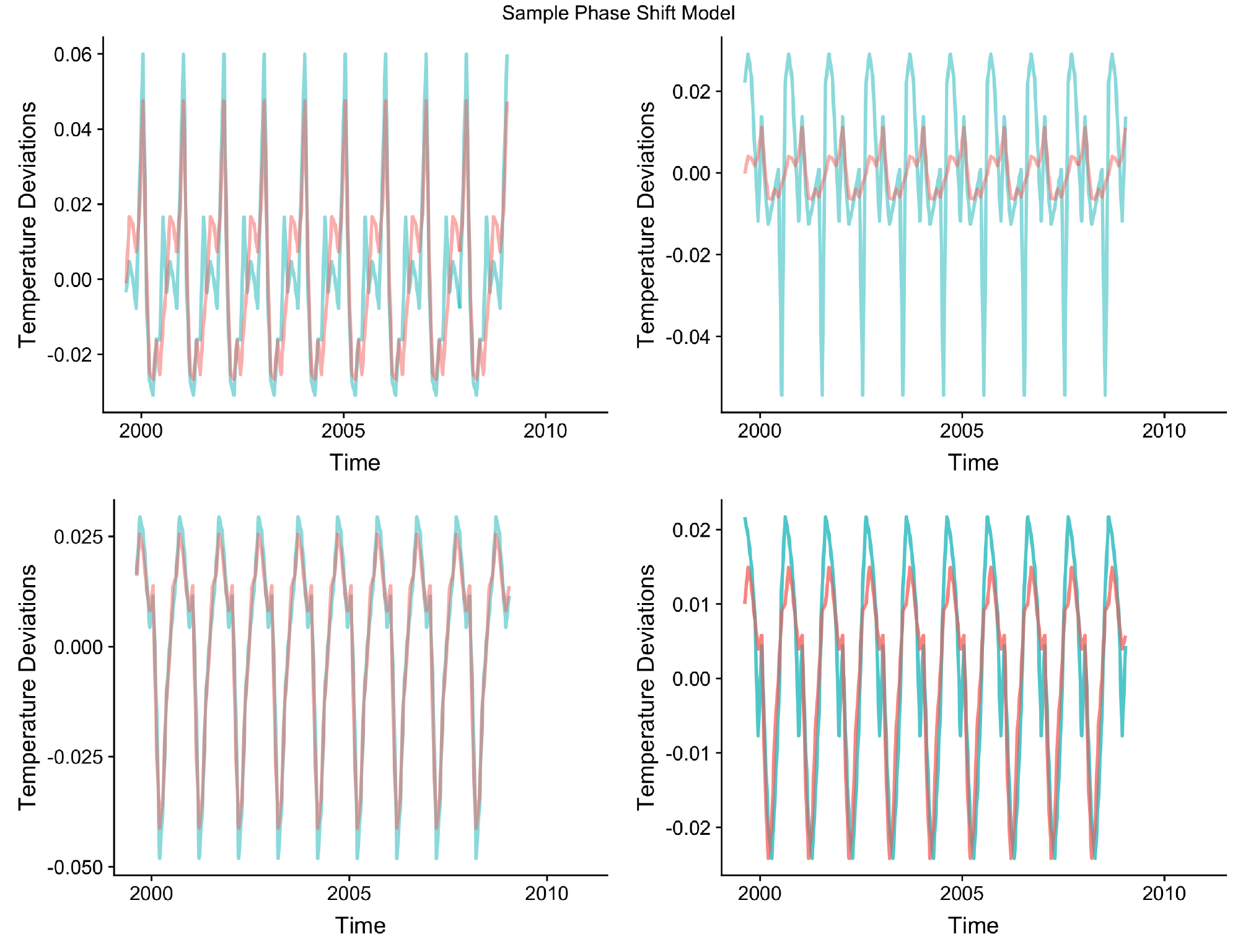 --- # Problems with the approach so far - Two parameters per location (for large problems that is a lot) -- - Doesn't "borrow strength" spatially + Local spatial differences not properly modeled -- - Add convolution kernels + Processes only defined at the "knots" + kernel then "spreads" the effects spatially + shape and distance of kernel estimated (but usually only a couple of parameters per knot) --- # Modeling SST with phase delay and kernel - Data is pre-computed season+cycle for 100 locations + (150W, 143.5W), (43.5N, 48N) + Relatively homogeneous area -- - Kernel's are pre-computed, not estimated + kernel grid inside obs grid, usually the opposite -- - Fit model only includes a seasonal term --- # Sample Results 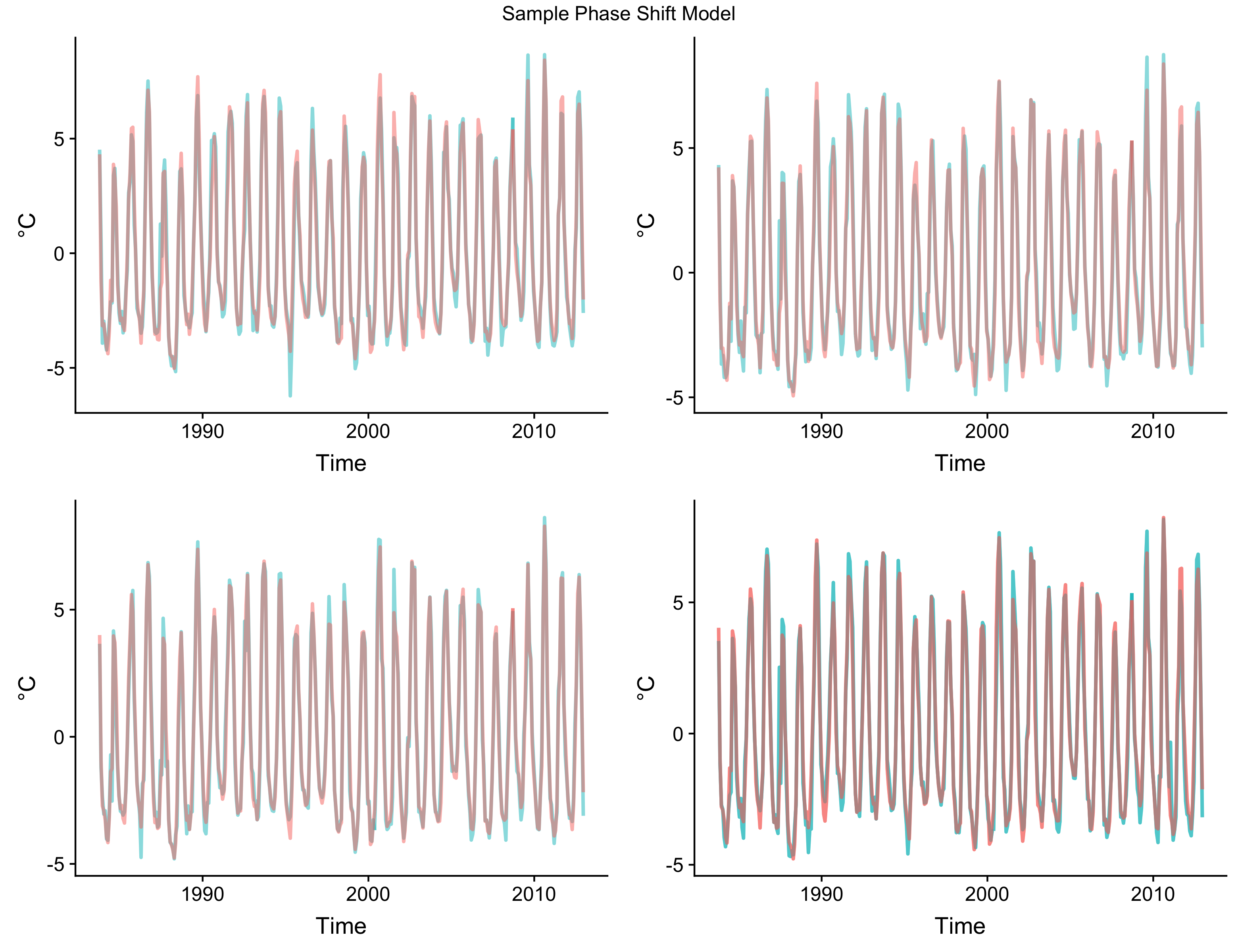 --- # Still to do - Estimate kernel parameters -- - "Good" kernel grid - particularly near-shore -- - Dealing with large problem size and missing data + Jungbacker-Koopman transformation + Univariate (one-at-a-time) processing + Likely will have to write special purpose code --- # Still to do - model the cross-relationship with tuna -- - Big Problems + Most locations have no obs. at most time periods + less sanguine than others that you can unconfound fleet behavior from fish behavior + particular since tuna fleet uses environmental cues to determine where to fish ---